Projects
Below are some of my side projects that are not directly related to my thesis. Titles are linked to repos/slide decks when available.
Disease Gene Classification
Description: In collaboration with Johns Hopkins University School of Medicine (Dr. Sandeep Wontakal), an expansive gene network was created, capturing diverse interactions of orthologous genes across 7 species. Gene properties and interactions were organized into a graph. Computational methods are being developed to leverage graph properties to probabilistically assign unclassified genes as disease related or non-disease related genes. In Progress
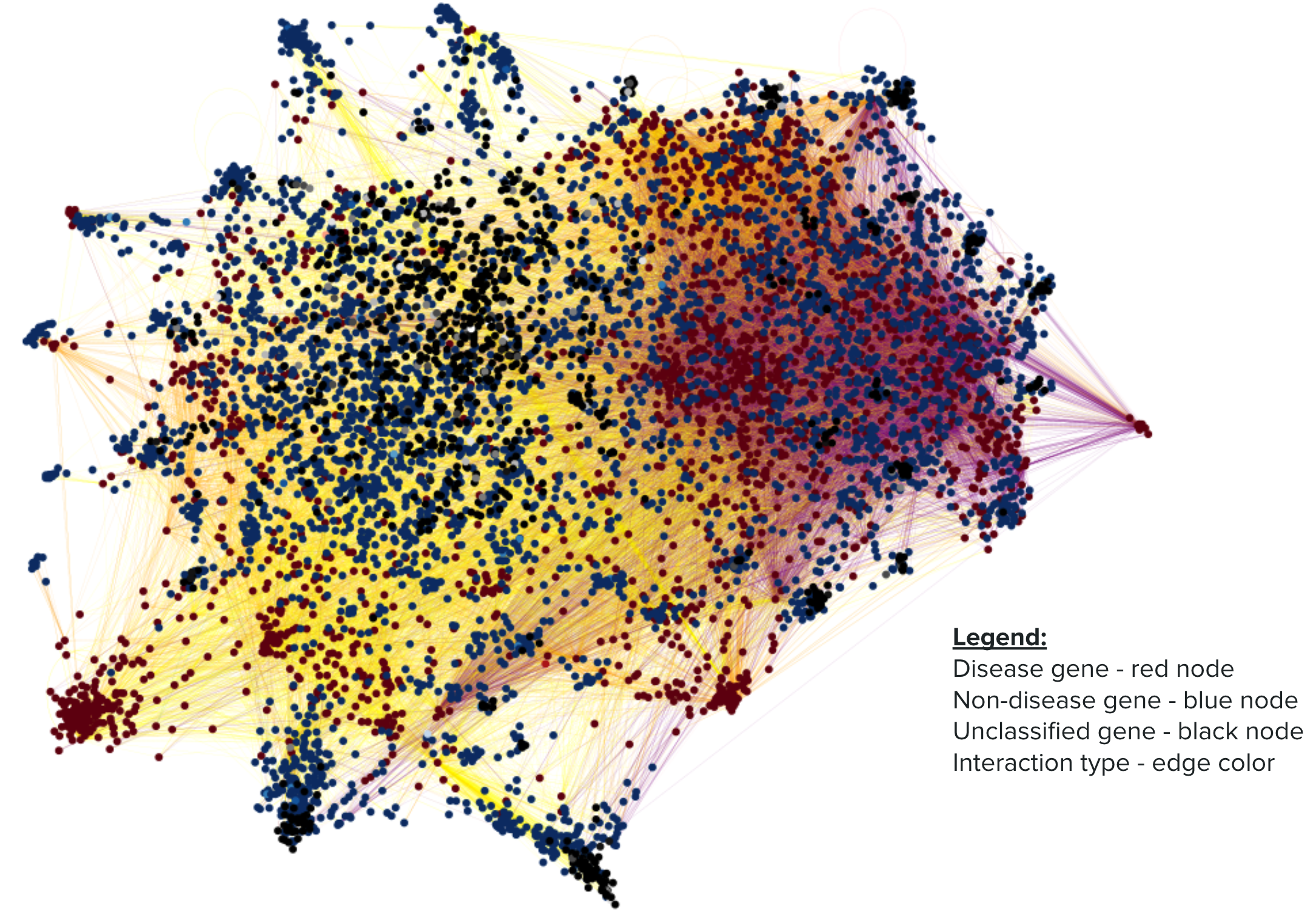
Figures: node2vec was applied to each gene in the graph to generate embeddings. The embeddings were then visualized using UMAP. This visualization represents 4000 nodes and 40,000 edges.
Generalizable Deep Learning for a Handwriting BCI Decoder
Description: The act of imagining handwriting generates neural activity patterns in the hand motor cortex area that can be decoded with BCIs to generate letters and words. Improving deep learning models to decode neural activity provides a promising avenue for restoring real-time communication for individuals suffering from paralysis and motor-related disorders. In this project, we trained CNN and RNN models to decode handwriting at high accuracy without daily recalibriation, building on research by Willet et al., 2021. This project was completed throughout the neuromatch academy deep learning course. 2024
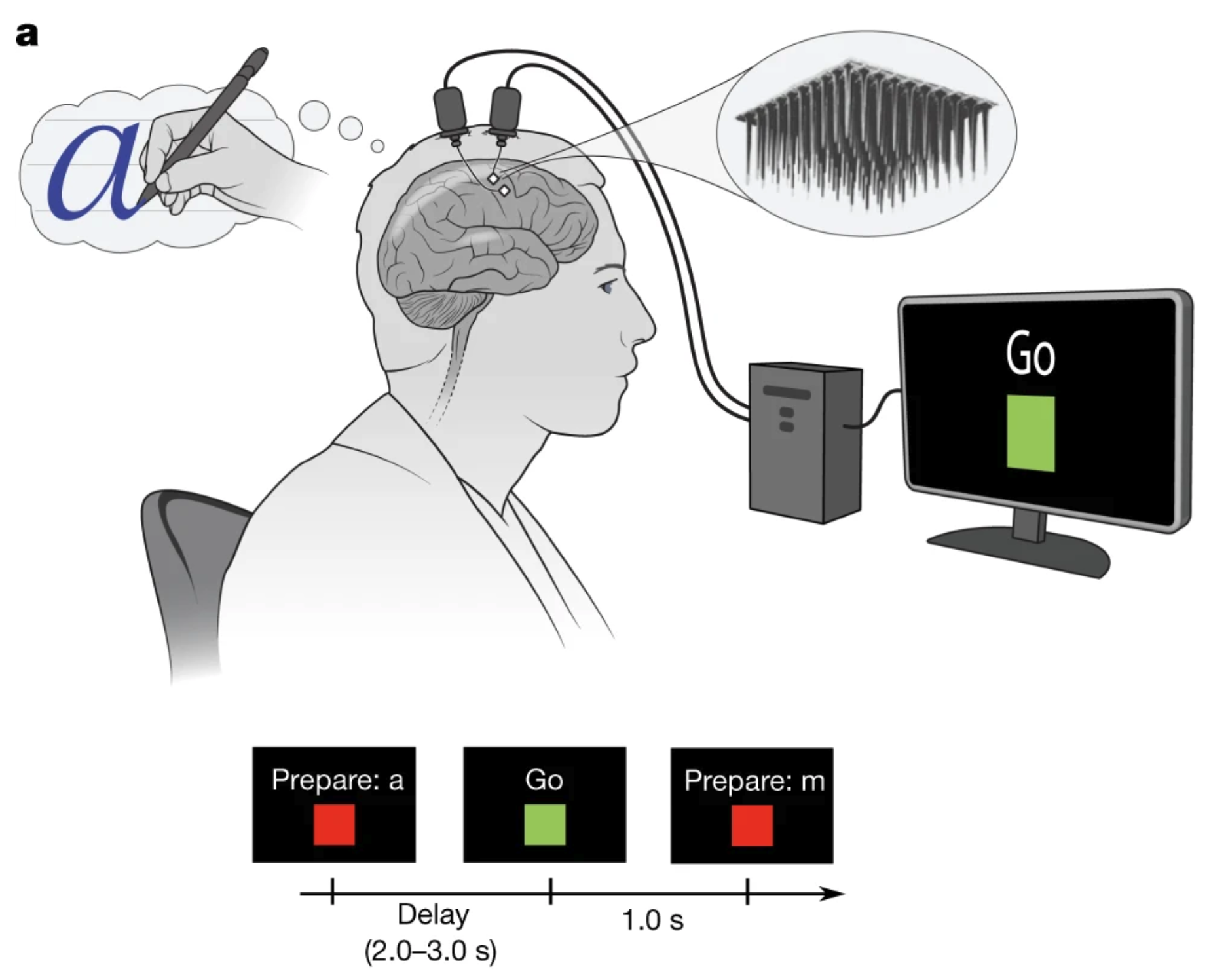
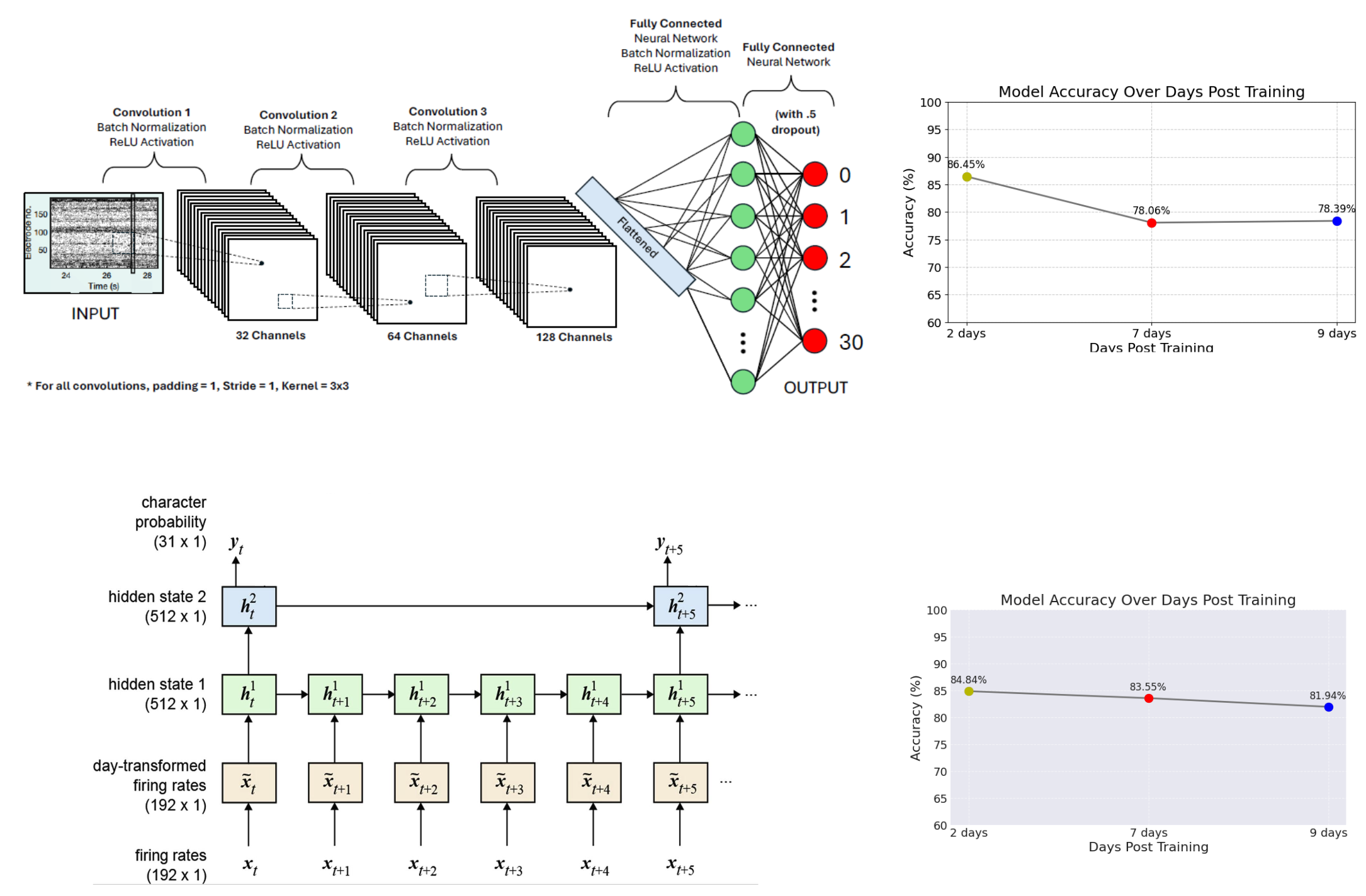
Figures: Top - experiment schematic showing microelectrode array capturing signals from motor cortex. Bottom - CNN and RNN architectures and days after training performance curves.
Intracortical Gene Expression across Primate Species
Description: In comparison to close evolutionary relatives, the human brain has distinct properties from a biological and behavioral standpoint. The present study aims to highlight differences in orthologous gene expression patterns between humans and chimpanzees. Focusing on two regions with central roles in cognition, the anterior cingulate cortex and cerebellar white matter, differential expression analysis identified sets of human-specific overexpressed genes in each region respectively. These gene expression patterns shed light on the evolutionary functional changes acquired in the human lineage beyond gene duplication events. 2024 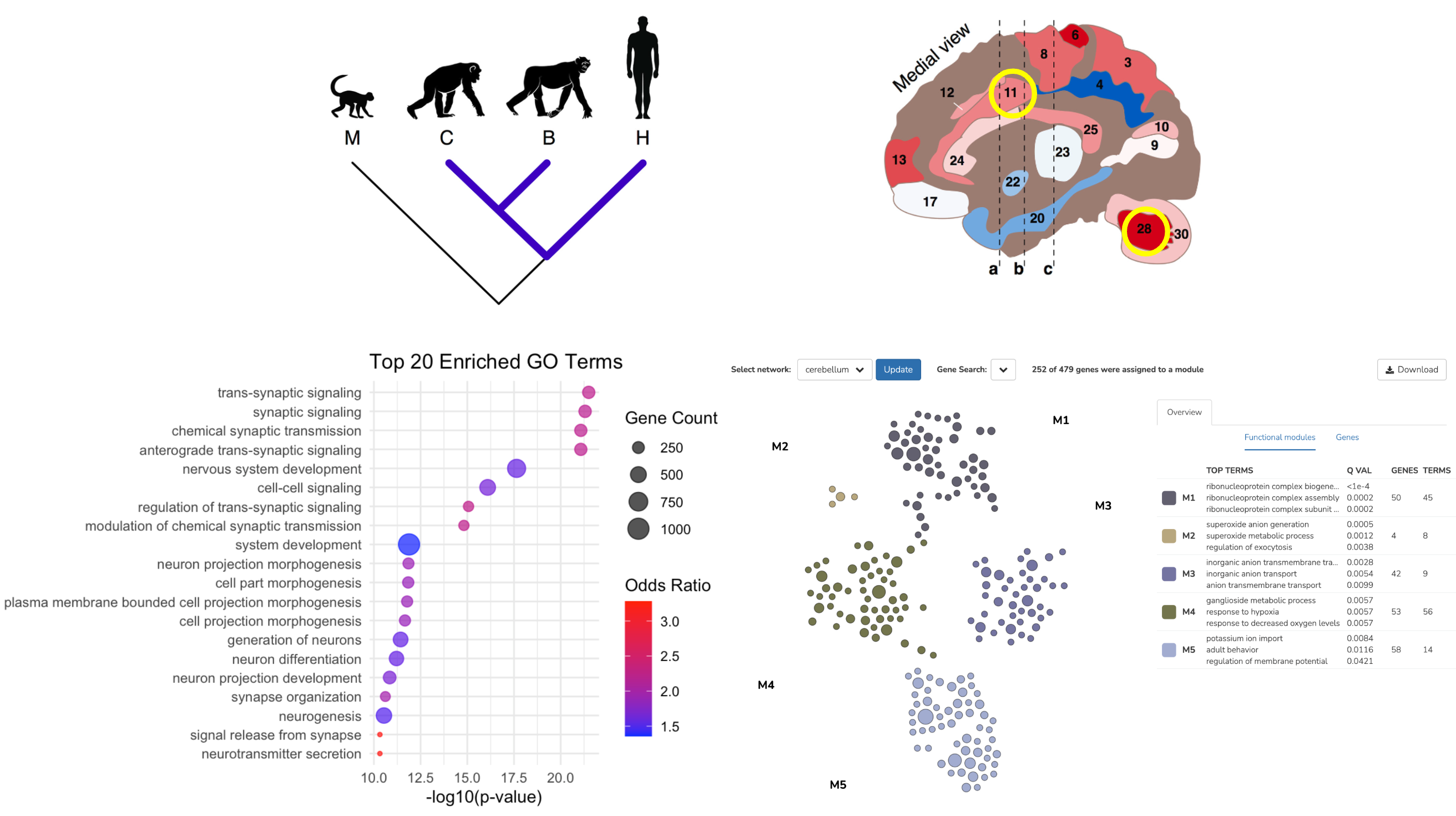
Figures: Top - phylogenetic tree schematic. Sagitall cross-section of brain highlighting the anterior cingulate cortex (11) and cerebellar white matter (28). Bottom - Enriched GO terms for differentlially expressed genes in the cerebellar white matter. Functional module plot created with humanbase.
Clinical Subtyping of Multiple Sclerosis with SuStaIn
Description: This project is in collaboration with Columbia University Medical Center (Dr. Victoria Leavitt). The objective of the project is to derive subtypes and subtype stages based solely on clinical data from a large longitudinal cohort of patients with Multiple Sclerosis (MS). We specifically focus on behavioral clinical data that can be recorded at clinical visits. To derive subtypes that differ both phenotypically and temporally, we combine two related methods - Subtype and Stage Inference with Temporal Event-Based Modeling. In this ongoing work, we have fit preliminary models to identify 3 subtypes that differ in their disease progression as well as demographic and neurophysiological features. The final model should be able to assign an unseen patient to a given subtype and accurately predict biomarker progression. In Progress
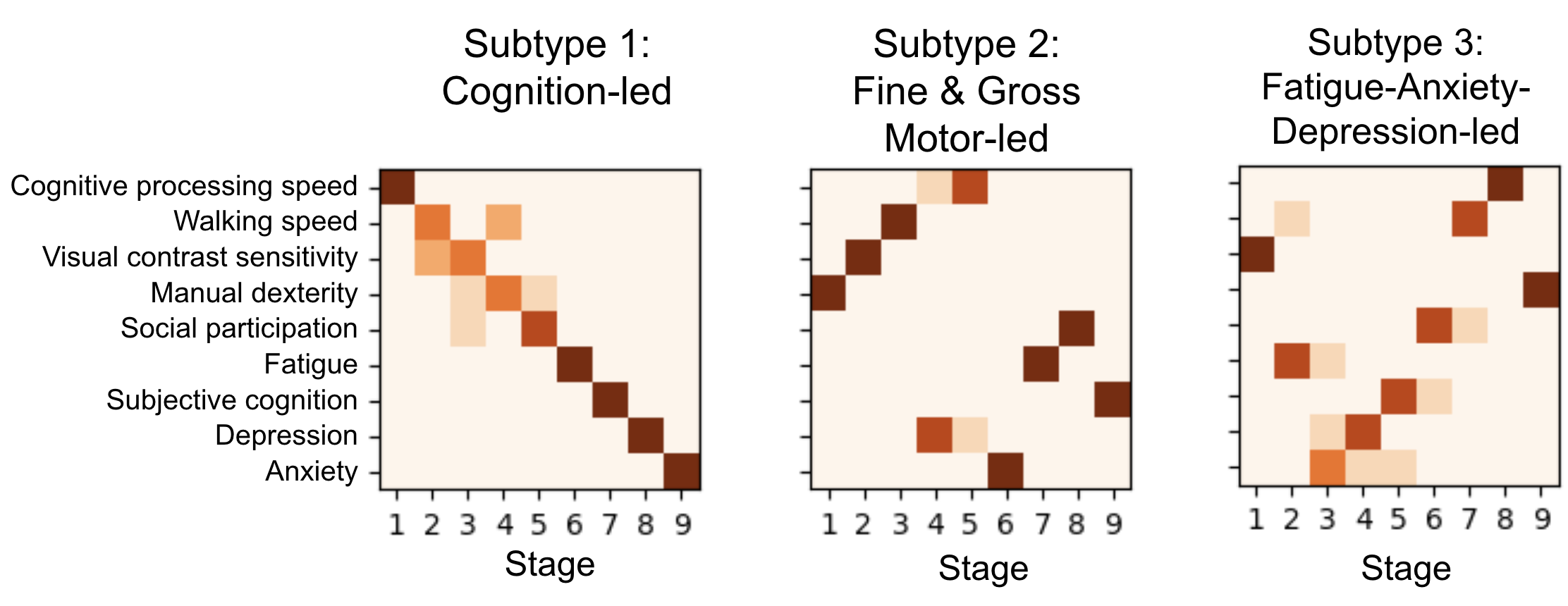
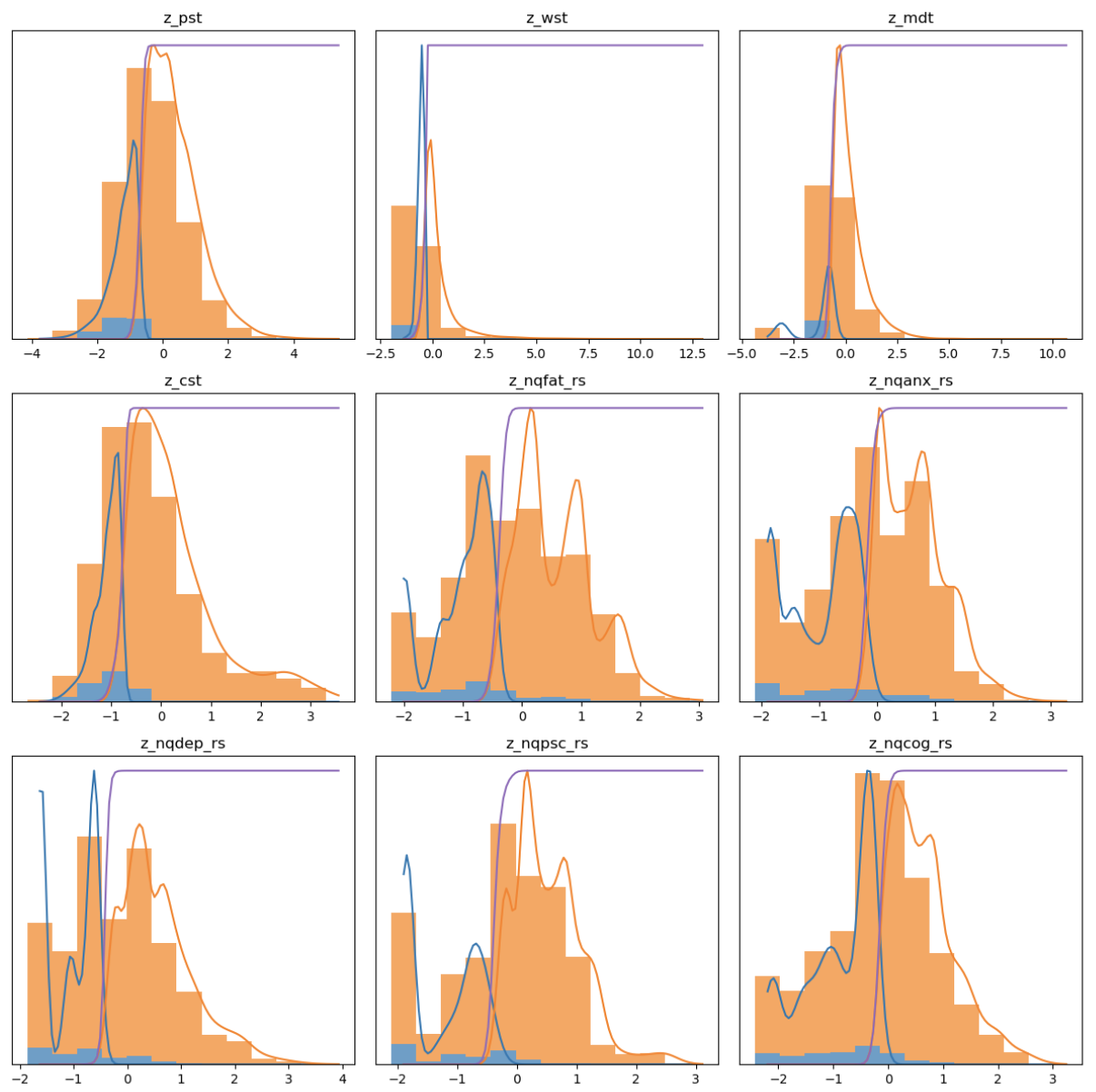
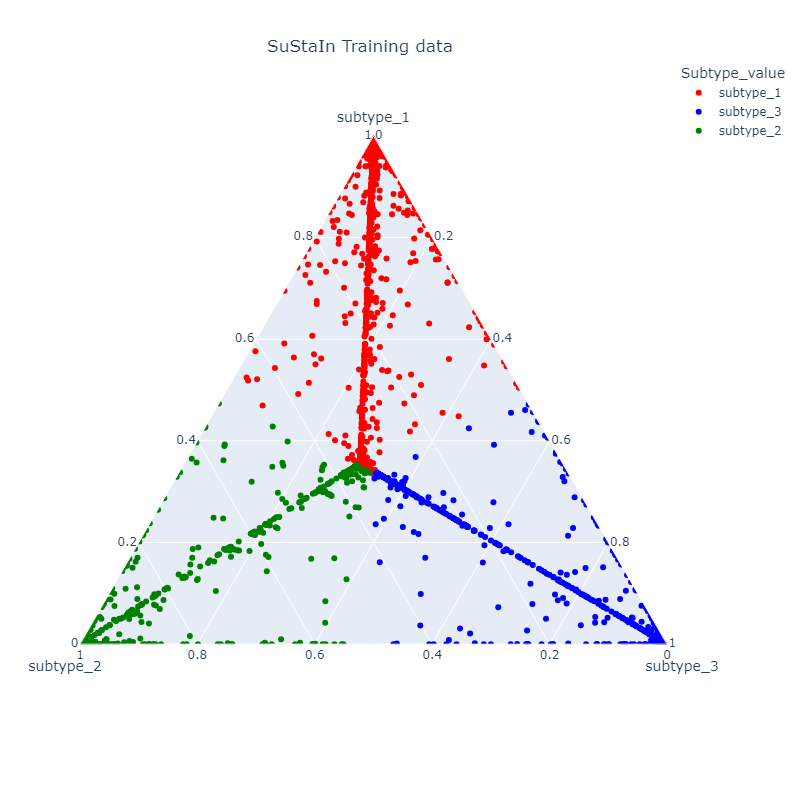
Figures: Top - derived subtypes from patient data showing order of clinical progression. Left - kernel density estimates of clinical features between patients and asymptomatic patients. Right - Each point corresponds to a single subject and the location in the triangle reflects confidence of assignment to each subtype.
NeuroAnalysis Project
Description: Applied various machine learning techniques (Support Vector Machines, Clustering, Multi-Dimensional
Scaling, Principal Component Analysis) and statistical analysis tools (Representational Similarity Analysis, correlational maps and matrices) to classify, predict, and examine neural data. 2020
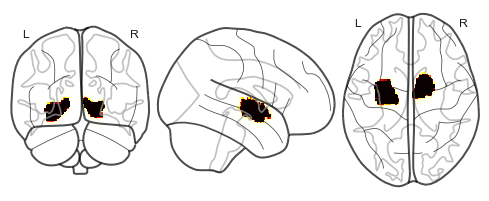
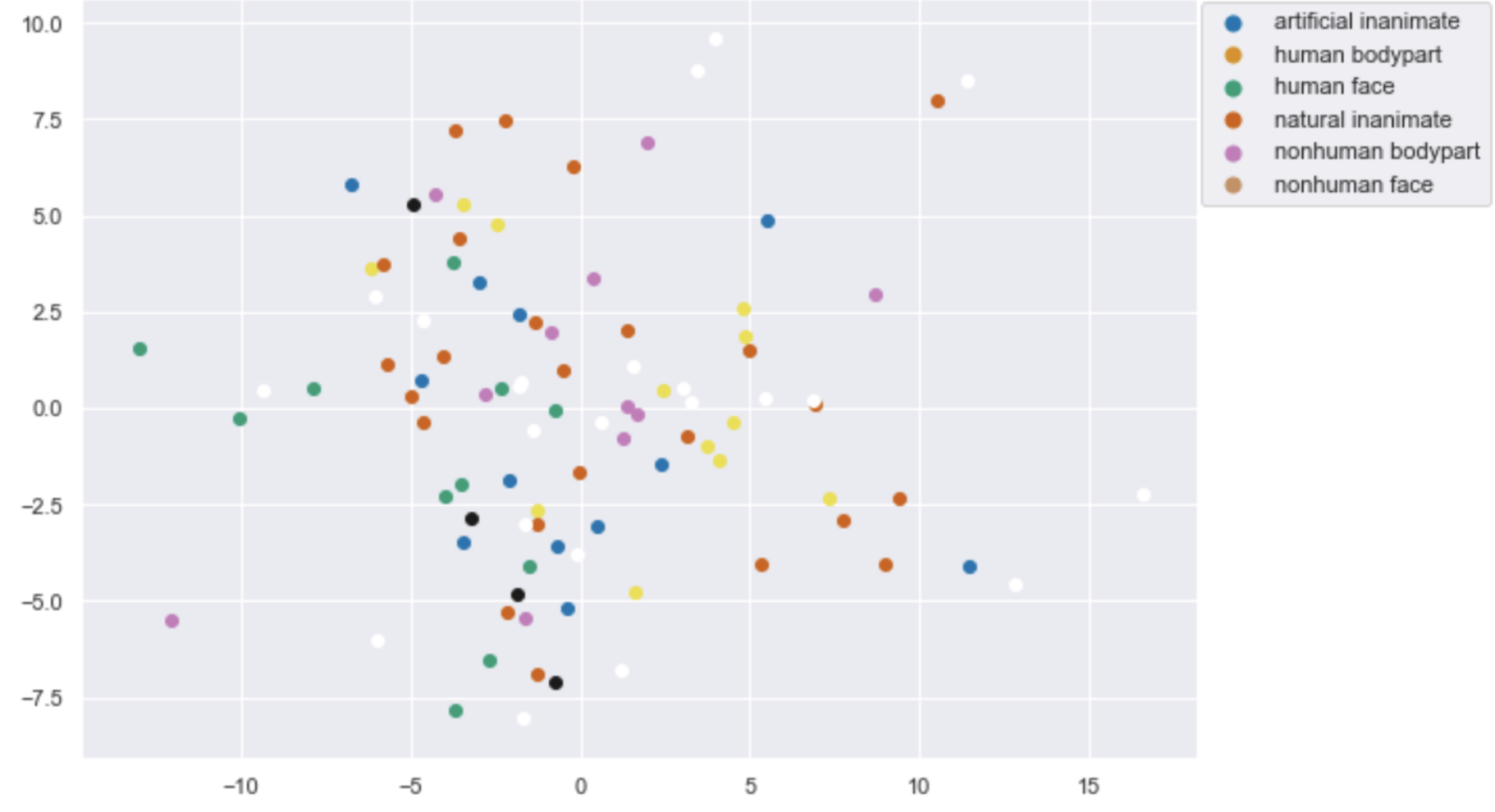
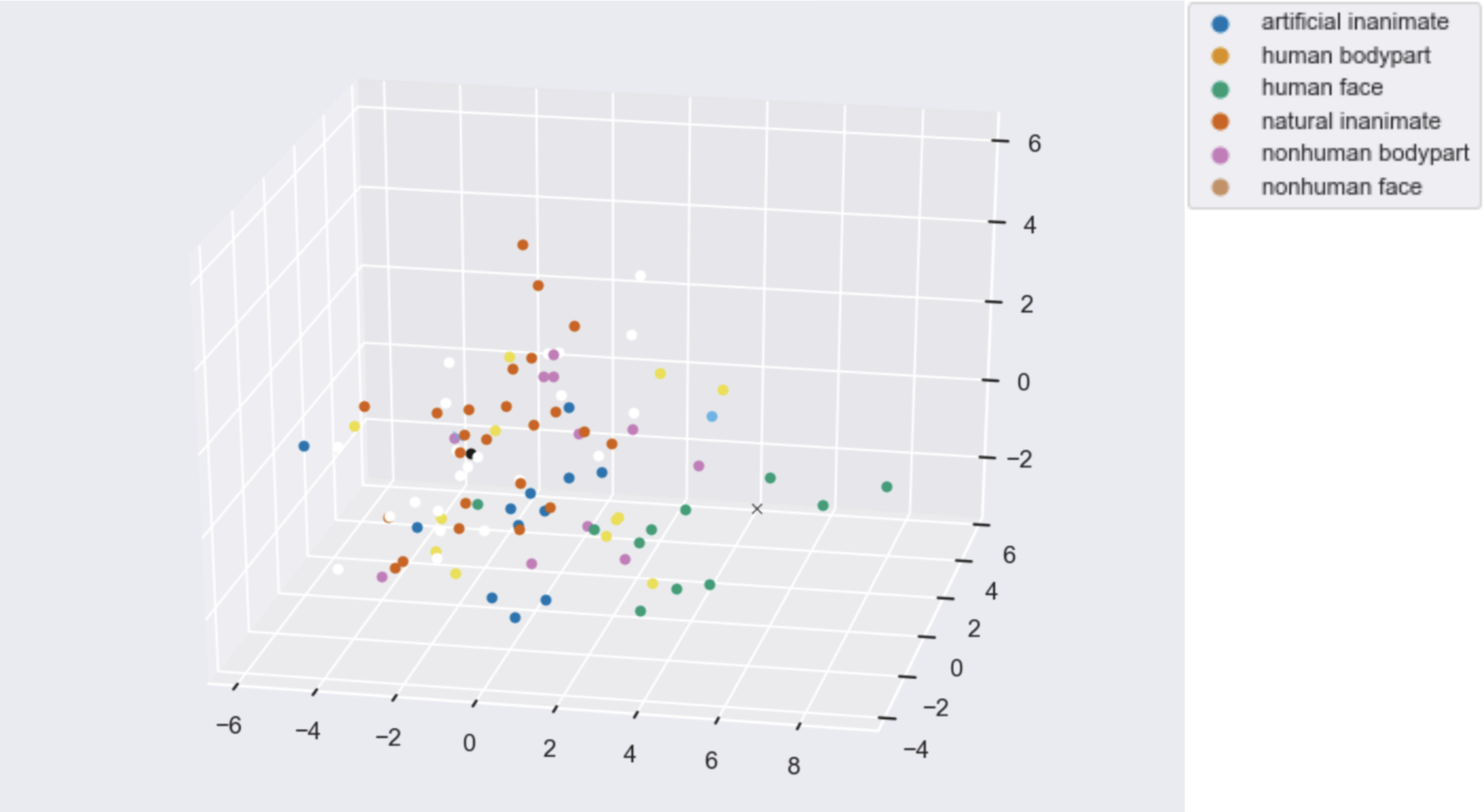
Figures: Left - Parahippocampal place area mask from which fMRI data was extracted while subjects viewed various items. Middle - 2D representation of fMRI data from PCA. Right - 3D representation of fMRI data from PCA.
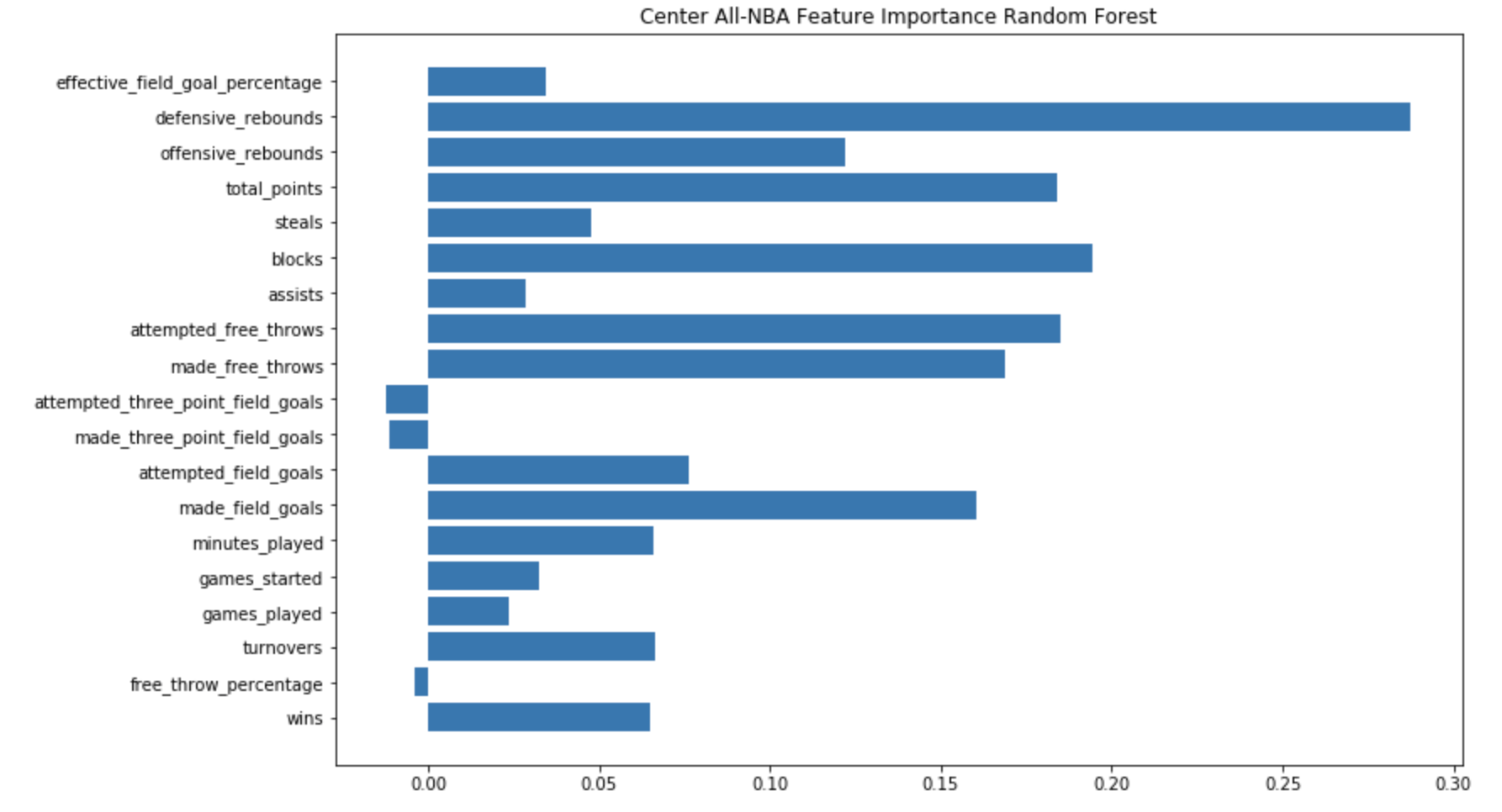
All-NBA Machine Learning
Description: Predicted the 2020 All-NBA team with 87% accuracy using a multi-layer perceptron neural net optimized and trained on 20 seasons of NBA statistics. 2019